The weekly litigation news digest is live. Subscribe now
Crispr-Based Genome Modification And Regulation - EP3138911
The patent EP3138911 was granted to Sigma Aldrich on Dec 5, 2018. The application was filed on Dec 5, 2013 under application number EP16183724A. The patent is currently recorded with a legal status of "Revoked".
EP3138911
- Application Number
- EP16183724A
- Filing Date
- Dec 5, 2013
- Status
- Revoked
- Nov 22, 2024
- Publication Date
- Dec 5, 2018
- External Links
- Slate, Register, Google Patents
Patent Summary
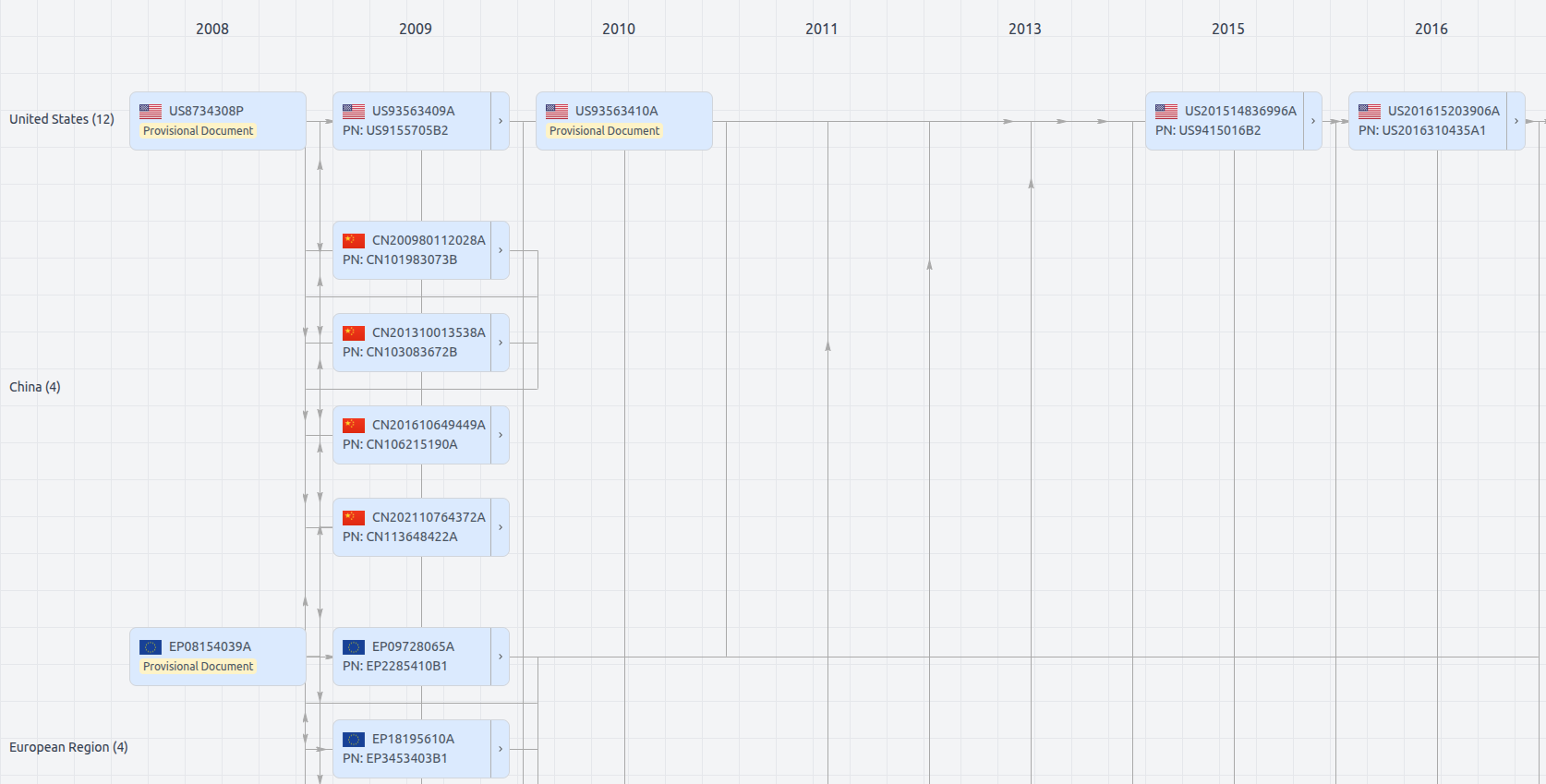
Patent Family
Patent Oppositions (9)
Patent oppositions filed by competitors challenge the validity of a granted patent. These oppositions are typically based on claims of prior art, lack of novelty, or non-obviousness. They are a key part of the process for determining a patent's strength and enforceability.
| Company | Opposition Date | Representative | Opposition Status |
|---|---|---|---|
| MATHYS & SQUIRE | Sep 5, 2019 | WILDING | ADMISSIBLE |
| POHLMAN | Sep 5, 2019 | POHLMAN | ADMISSIBLE |
| COOLEY UK | Sep 5, 2019 | COOLEY UK | WITHDRAWN |
| BASF | Sep 4, 2019 | EISENFUHR SPEISER | ADMISSIBLE |
| PATENT BOUTIQUE | Sep 4, 2019 | PATENT BOUTIQUE | ADMISSIBLE |
| BAYER BAYER | Jun 18, 2019 | COHAUSZ & FLORACK | ADMISSIBLE |
| VOSSIUS & PARTNER PATENTANWALTE RECHTSANWALTE MBB | Feb 22, 2019 | VOSSIUS & PARTNER PATENTANWALTE RECHTSANWALTE MBB | ADMISSIBLE |
| GRUND IPG PATENTANWALTE UND SOLICITOR PARTG MBB | Jan 30, 2019 | GRUND | ADMISSIBLE |
| SCHLICH | Dec 6, 2018 | SCHLICH | ADMISSIBLE |
Patent Citations (17) New
Patent citations refer to prior patents cited during different phases such as opposition or international search.
| Citation Phase | Publication Number |
|---|---|
| OPPOSITION | EP2800811 |
| OPPOSITION | EP2912175 |
| OPPOSITION | EP3138910 |
| OPPOSITION | EP3241902 |
| OPPOSITION | EP3401400 |
| OPPOSITION | US2005220796 |
| OPPOSITION | US2010076057 |
| OPPOSITION | WO2011146121 |
| OPPOSITION | WO2013176772 |
| OPPOSITION | WO2014065596 |
| OPPOSITION | WO2014089290 |
| OPPOSITION | WO2014093595 |
| SEARCH | US2010076057 |
| SEARCH | WO2013176772 |
| SEARCH | WO2014065596 |
| SEARCH | WO2014093655 |
| SEARCH | WO2014099744 |
Dossier Documents
The dossier documents provide a comprehensive record of the patent's prosecution history - including filings, correspondence, and decisions made by patent offices - and are crucial for understanding the patent's legal journey and any challenges it may have faced during examination.
Date
Description
Get instant alerts for new documents
Nov 29, 2024
Nov 29, 2024
Nov 22, 2024
Refund of fees
Appeal
Nov 20, 2024
Nov 20, 2024
Nov 20, 2024
Nov 20, 2024
Nov 20, 2024
Nov 20, 2024
Nov 20, 2024
Nov 20, 2024
Nov 20, 2024
Nov 19, 2024
Nov 19, 2024
Nov 19, 2024
Nov 19, 2024
Nov 19, 2024
Nov 19, 2024
Nov 19, 2024
Nov 19, 2024
Nov 19, 2024
Nov 19, 2024
Nov 7, 2024
Nov 6, 2024
(Electronic) Receipt
Appeal
Nov 6, 2024
Nov 6, 2024
Oct 24, 2024
Oct 23, 2024
(Electronic) Receipt
Appeal
Oct 23, 2024
Oct 23, 2024
Oct 23, 2024
Oct 21, 2024
Oct 18, 2024
(Electronic) Receipt
Appeal
Oct 18, 2024
Oct 18, 2024
Oct 16, 2024
(Electronic) Receipt
Appeal
Oct 16, 2024
Oct 16, 2024
Oct 16, 2024
Oct 16, 2024
Oct 16, 2024
Oct 16, 2024
Oct 16, 2024
Oct 16, 2024
Oct 16, 2024
Oct 16, 2024
Oct 16, 2024
Oct 16, 2024
Oct 15, 2024
Oct 15, 2024
Oct 15, 2024
Oct 11, 2024
(Electronic) Receipt
Appeal
Oct 11, 2024
Oct 11, 2024
Oct 10, 2024
(Electronic) Receipt
Appeal
Oct 10, 2024
Oct 10, 2024
Oct 10, 2024
Oct 10, 2024
Oct 10, 2024
Oct 10, 2024
Oct 10, 2024
Oct 10, 2024
Oct 10, 2024
Oct 10, 2024
Oct 9, 2024
(Electronic) Receipt
Appeal
Oct 9, 2024
Oct 9, 2024
Oct 8, 2024
(Electronic) Receipt
Appeal
Oct 8, 2024
Oct 8, 2024
Sep 17, 2024
Sep 17, 2024
Sep 12, 2024
Sep 12, 2024
Sep 11, 2024
(Electronic) Receipt
Appeal
Sep 11, 2024
(Electronic) Receipt
Appeal
Sep 11, 2024
(Electronic) Receipt
Appeal
Sep 11, 2024
Sep 11, 2024
Sep 11, 2024
Sep 11, 2024
Sep 11, 2024
Sep 11, 2024
Sep 4, 2024
Sep 4, 2024
Sep 4, 2024
Sep 4, 2024
Sep 4, 2024
Sep 4, 2024
Sep 4, 2024
Sep 4, 2024
Aug 29, 2024
(Electronic) Receipt
Appeal
Aug 29, 2024
Aug 29, 2024
Aug 9, 2024
Aug 6, 2024
(Electronic) Receipt
Appeal
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 6, 2024
Aug 17, 2023
Aug 10, 2023
(Electronic) Receipt
Appeal
Aug 10, 2023
Aug 10, 2023
Aug 10, 2023
Jul 14, 2023
(Electronic) Receipt
Appeal
Jul 14, 2023
Advice of delivery
Appeal
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 14, 2023
Jul 11, 2023
Jul 4, 2023
Jun 29, 2023
Jun 28, 2023
(Electronic) Receipt
Appeal
Jun 28, 2023
Jun 28, 2023
Dec 7, 2022
Dec 2, 2022
Dec 2, 2022
Dec 1, 2022
Dec 1, 2022
Nov 30, 2022
Nov 30, 2022
Nov 25, 2022
(Electronic) Receipt
Appeal
Nov 25, 2022
(Electronic) Receipt
Appeal
Nov 25, 2022
(Electronic) Receipt
Appeal
Nov 25, 2022
(Electronic) Receipt
Appeal
Nov 25, 2022
(Electronic) Receipt
Appeal
Nov 25, 2022
(Electronic) Receipt
Appeal
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Nov 25, 2022
Non-scannable object
Appeal
Nov 25, 2022
Non-scannable object
Appeal
Nov 25, 2022
Reply to appeal
Appeal
Nov 25, 2022
Reply to appeal
Appeal
Nov 25, 2022
Reply to appeal
Appeal
Nov 25, 2022
Reply to appeal
Appeal
Nov 25, 2022
Reply to appeal
Appeal
Nov 25, 2022
Reply to appeal
Appeal
Nov 25, 2022
Reply to appeal
Appeal
Nov 25, 2022
Reply to appeal
Appeal
Nov 23, 2022
Nov 18, 2022
Nov 17, 2022
(Electronic) Receipt
Appeal
Nov 17, 2022
Nov 17, 2022
Reply to appeal
Appeal
Nov 17, 2022
Reply to appeal
Appeal
Nov 15, 2022
(Electronic) Receipt
Appeal
Nov 15, 2022
Nov 15, 2022
Nov 15, 2022
Nov 15, 2022
Nov 15, 2022
Nov 15, 2022
Reply to appeal
Appeal
Aug 18, 2022
Aug 8, 2022
Aug 3, 2022
(Electronic) Receipt
Appeal
Aug 3, 2022
Aug 3, 2022
Aug 3, 2022
Jul 19, 2022
Jul 15, 2022
Jul 14, 2022
(Electronic) Receipt
Appeal
Jul 14, 2022
Jul 14, 2022
Jul 14, 2022
Jul 11, 2022
(Electronic) Receipt
Appeal
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jul 11, 2022
Jun 10, 2022
Jun 10, 2022
Jun 10, 2022
Jun 10, 2022
Jun 10, 2022
Jun 10, 2022
May 9, 2022
Apr 29, 2022
(Electronic) Receipt
Appeal
Apr 29, 2022
Apr 29, 2022
Notice of appeal
Appeal
Mar 15, 2022
Mar 8, 2022
Mar 8, 2022
Mar 8, 2022
Mar 4, 2022
Mar 2, 2022
Mar 2, 2022
Mar 2, 2022
Mar 2, 2022
Mar 2, 2022
Mar 2, 2022
Mar 2, 2022
Mar 2, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Means of redress
OPPO
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Mar 1, 2022
Jan 5, 2022
Jan 5, 2022
Jan 5, 2022
Jan 5, 2022
Jan 5, 2022
Jan 5, 2022
Jan 5, 2022
Jan 5, 2022
Jan 5, 2022
Jan 5, 2022
Dec 21, 2021
Dec 21, 2021
Dec 21, 2021
Nov 9, 2021
Nov 8, 2021
Nov 8, 2021
Nov 8, 2021
Nov 8, 2021
Nov 8, 2021
Nov 8, 2021
Nov 8, 2021
Nov 8, 2021
Nov 8, 2021
Nov 5, 2021
Nov 5, 2021
Nov 5, 2021
Nov 5, 2021
Nov 5, 2021
Nov 5, 2021
Nov 5, 2021
Nov 5, 2021
Nov 5, 2021
Nov 4, 2021
Nov 4, 2021
Nov 4, 2021
Nov 4, 2021
Nov 4, 2021
Nov 4, 2021
Nov 4, 2021
Nov 4, 2021
Nov 4, 2021
Nov 2, 2021
Nov 2, 2021
Nov 2, 2021
Nov 2, 2021
Nov 2, 2021
Nov 2, 2021
Nov 2, 2021
Nov 2, 2021
Nov 2, 2021
Nov 2, 2021
Nov 2, 2021
Oct 28, 2021
Oct 28, 2021
Oct 28, 2021
Oct 28, 2021
Oct 28, 2021
Oct 27, 2021
Oct 27, 2021
Oct 27, 2021
Oct 27, 2021
Oct 27, 2021
Oct 27, 2021
Oct 27, 2021
Oct 27, 2021
Oct 27, 2021
Oct 22, 2021
Oct 22, 2021
Oct 22, 2021
Oct 22, 2021
Oct 7, 2021
Oct 7, 2021
Oct 7, 2021
Oct 7, 2021
Oct 7, 2021
Oct 7, 2021
Oct 7, 2021
Oct 7, 2021
Oct 7, 2021
Oct 5, 2021
Oct 5, 2021
Oct 5, 2021
Oct 5, 2021
Oct 5, 2021
Oct 5, 2021
Oct 5, 2021
Oct 5, 2021
Oct 5, 2021
Oct 4, 2021
Oct 4, 2021
Oct 4, 2021
Oct 4, 2021
Oct 4, 2021
Oct 4, 2021
Oct 4, 2021
Oct 4, 2021
Oct 4, 2021
Oct 4, 2021
Oct 4, 2021
Oct 1, 2021
Oct 1, 2021
Oct 1, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 30, 2021
Sep 14, 2021
Sep 14, 2021
Sep 14, 2021
Sep 14, 2021
Sep 14, 2021
Sep 14, 2021
Sep 14, 2021
Sep 14, 2021
Sep 14, 2021
Sep 14, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 8, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 7, 2021
Sep 2, 2021
Sep 2, 2021
Sep 2, 2021
Sep 2, 2021
Sep 2, 2021
Sep 2, 2021
Sep 2, 2021
Sep 2, 2021
Sep 2, 2021
Aug 26, 2021
Aug 26, 2021
Aug 26, 2021
Aug 24, 2021
Aug 24, 2021
Aug 24, 2021
Aug 24, 2021
Aug 24, 2021
Aug 24, 2021
Aug 24, 2021
Aug 24, 2021
Aug 24, 2021
Aug 18, 2021
Aug 18, 2021
Aug 18, 2021
Aug 4, 2021
Jul 30, 2021
Jul 30, 2021
Jul 30, 2021
Jul 30, 2021
Jul 30, 2021
Jul 30, 2021
Jul 30, 2021
Jul 30, 2021
Jul 30, 2021
Jul 27, 2021
Jul 27, 2021
Jul 27, 2021
Jul 27, 2021
Jul 27, 2021
Jul 27, 2021
Jul 27, 2021
Jul 21, 2021
Jul 21, 2021
Jul 21, 2021
Jul 21, 2021
Jul 21, 2021
Jul 21, 2021
Jul 21, 2021
Jul 21, 2021
Jul 21, 2021
Jul 15, 2021
Jul 15, 2021
Jul 15, 2021
Jul 9, 2021
Jul 9, 2021
Jul 9, 2021
Jul 9, 2021
Jul 9, 2021
Jul 9, 2021
Jul 9, 2021
Jul 9, 2021
Jul 9, 2021
Jul 9, 2021
May 17, 2021
May 17, 2021
Mar 5, 2021
Mar 5, 2021
Mar 5, 2021
Mar 5, 2021
Mar 5, 2021
Mar 5, 2021
Mar 5, 2021
Mar 5, 2021
Mar 5, 2021
Feb 26, 2021
Feb 26, 2021
Feb 26, 2021
Feb 26, 2021
Feb 26, 2021
Feb 26, 2021
Feb 15, 2021
Jan 21, 2021
Jan 21, 2021
Jan 21, 2021
Nov 11, 2020
Nov 11, 2020
Nov 11, 2020
Nov 6, 2020
Nov 6, 2020
Nov 6, 2020
Nov 4, 2020
Nov 3, 2020
Nov 3, 2020
Nov 3, 2020
Nov 2, 2020
Nov 2, 2020
Nov 2, 2020
Nov 2, 2020
Oct 30, 2020
Oct 29, 2020
Oct 29, 2020
Oct 29, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Oct 28, 2020
Sep 4, 2020
Communication of amended entries
Search/Exam
Aug 3, 2020
Jul 22, 2020
Jul 22, 2020
Jul 22, 2020
Jul 22, 2020
Jul 22, 2020
Jul 22, 2020
Jul 22, 2020
Jul 22, 2020
Jul 22, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 16, 2020
Jul 9, 2020
Request concerning representative opponent
Search/Exam
Jun 9, 2020
Jun 9, 2020
Jun 9, 2020
Jun 9, 2020
Jun 9, 2020
Jun 9, 2020
Jun 9, 2020
Jun 9, 2020
Jun 9, 2020
Jun 9, 2020
May 26, 2020
May 26, 2020
May 26, 2020
May 26, 2020
May 26, 2020
May 26, 2020
May 26, 2020
May 26, 2020
May 26, 2020
May 19, 2020
May 19, 2020
May 19, 2020
May 18, 2020
May 18, 2020
May 18, 2020
May 18, 2020
Apr 14, 2020
Apr 14, 2020
Apr 14, 2020
Apr 14, 2020
Apr 14, 2020
Apr 14, 2020
Apr 14, 2020
Apr 14, 2020
Apr 14, 2020
Apr 14, 2020
Apr 1, 2020
Mar 11, 2020
Mar 11, 2020
Mar 11, 2020
Mar 6, 2020
Mar 6, 2020
Mar 6, 2020
Mar 6, 2020
Mar 6, 2020
Mar 6, 2020
Mar 6, 2020
Mar 6, 2020
Mar 6, 2020
Mar 6, 2020
Feb 24, 2020
Feb 24, 2020
Feb 24, 2020
Sep 19, 2019
Sep 19, 2019
Sep 19, 2019
Sep 12, 2019
Sep 12, 2019
Sep 12, 2019
Sep 11, 2019
Sep 11, 2019
Sep 11, 2019
Sep 11, 2019
Sep 10, 2019
Sep 10, 2019
Sep 9, 2019
Sep 9, 2019
Sep 9, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 6, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 5, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Sep 4, 2019
Aug 6, 2019
Aug 6, 2019
Aug 6, 2019
Aug 6, 2019
Aug 6, 2019
Jun 27, 2019
Jun 18, 2019
Jun 18, 2019
Jun 18, 2019
Feb 28, 2019
Feb 22, 2019
Feb 22, 2019
Feb 22, 2019
Feb 4, 2019
Jan 30, 2019
Jan 30, 2019
Jan 30, 2019
Dec 17, 2018
Dec 14, 2018
Dec 6, 2018
Dec 6, 2018
Dec 6, 2018
Nov 8, 2018
Decision to grant a European patent
Search/Exam
Oct 16, 2018
(Electronic) Receipt
Search/Exam
Oct 16, 2018
Filing of the translations of the claims
Search/Exam
Oct 16, 2018
French translation of claims
Search/Exam
Oct 16, 2018
German translation of the claims
Search/Exam
Oct 16, 2018
Letter accompanying subsequently filed items
Search/Exam
Oct 11, 2018
Oct 11, 2018
Oct 11, 2018
Oct 11, 2018
Intention to grant (signatures)
Search/Exam
Oct 11, 2018
Text intended for grant (clean copy)
Search/Exam
Oct 11, 2018
Text intended for grant (sequence listing)
Search/Exam
Oct 11, 2018
Aug 6, 2018
(Electronic) Receipt
Search/Exam
Aug 6, 2018
Amended claims with annotations
Search/Exam
Aug 6, 2018
Amended description with annotations
Search/Exam
Aug 6, 2018
Claims
Search/Exam
Aug 6, 2018
Description
Search/Exam
Aug 6, 2018
Letter accompanying subsequently filed items
Search/Exam
Aug 6, 2018
Matter concerning the application
Search/Exam
Jul 26, 2018
Jul 19, 2018
Observations by third parties
Search/Exam
Jul 5, 2018
Jul 2, 2018
(Electronic) Receipt
Search/Exam
Jul 2, 2018
Amended claims with annotations
Search/Exam
Jul 2, 2018
Amended description with annotations
Search/Exam
Jul 2, 2018
Claims
Search/Exam
Jul 2, 2018
Description
Search/Exam
Jul 2, 2018
Letter accompanying subsequently filed items
Search/Exam
Jul 2, 2018
Jun 29, 2018
Observations by third parties
Search/Exam
Jun 25, 2018
Annex to the communication
Search/Exam
Jun 25, 2018
Jun 25, 2018
Jun 5, 2018
May 31, 2018
Apr 6, 2018
(Electronic) Receipt
Search/Exam
Apr 6, 2018
Amended claims with annotations
Search/Exam
Apr 6, 2018
Amended description with annotations
Search/Exam
Apr 6, 2018
Claims
Search/Exam
Apr 6, 2018
Description
Search/Exam
Apr 6, 2018
Drawings
Search/Exam
Apr 6, 2018
Letter accompanying subsequently filed items
Search/Exam
Apr 6, 2018
Apr 6, 2018
Apr 6, 2018
Jan 15, 2018
Dec 22, 2017
Citation filed by a third party
Search/Exam
Dec 22, 2017
Citation filed by a third party
Search/Exam
Dec 22, 2017
Citation filed by a third party
Search/Exam
Dec 22, 2017
Citation filed by a third party
Search/Exam
Dec 22, 2017
Non-patent literature filed by a third party
Search/Exam
Dec 22, 2017
Non-patent literature filed by a third party
Search/Exam
Dec 22, 2017
Non-patent literature filed by a third party
Search/Exam
Dec 22, 2017
Observations by third parties
Search/Exam
Dec 22, 2017
Observations by third parties
Search/Exam
Dec 22, 2017
Observations by third parties
Search/Exam
Dec 22, 2017
Observations by third parties
Search/Exam
Dec 22, 2017
Observations by third parties
Search/Exam
Dec 22, 2017
Observations by third parties
Search/Exam
Dec 22, 2017
Observations by third parties
Search/Exam
Dec 22, 2017
Observations by third parties
Search/Exam
Dec 22, 2017
Observations by third parties
Search/Exam
Dec 22, 2017
Observations by third parties
Search/Exam
Dec 22, 2017
Observations by third parties
Search/Exam
Dec 22, 2017
Patent document filed by a Third party
Search/Exam
Dec 22, 2017
Patent document filed by a Third party
Search/Exam
Dec 22, 2017
Patent document filed by a Third party
Search/Exam
Dec 22, 2017
Patent document filed by a Third party
Search/Exam
Dec 22, 2017
Patent document filed by a Third party
Search/Exam
Dec 22, 2017
Patent document filed by a Third party
Search/Exam
Dec 22, 2017
Patent document filed by a Third party
Search/Exam
Dec 22, 2017
Patent document filed by a Third party
Search/Exam
Dec 14, 2017
Dec 11, 2017
Citation filed by a third party
Search/Exam
Dec 11, 2017
Citation filed by a third party
Search/Exam
Dec 11, 2017
Citation filed by a third party
Search/Exam
Dec 11, 2017
Citation filed by a third party
Search/Exam
Dec 11, 2017
Citation filed by a third party
Search/Exam
Dec 11, 2017
Non-patent literature filed by a third party
Search/Exam
Dec 11, 2017
Non-patent literature filed by a third party
Search/Exam
Dec 11, 2017
Non-patent literature filed by a third party
Search/Exam
Dec 11, 2017
Non-patent literature filed by a third party
Search/Exam
Dec 11, 2017
Non-patent literature filed by a third party
Search/Exam
Dec 11, 2017
Non-patent literature filed by a third party
Search/Exam
Dec 11, 2017
Observations by third parties
Search/Exam
Dec 11, 2017
Observations by third parties
Search/Exam
Dec 11, 2017
Observations by third parties
Search/Exam
Nov 29, 2017
Annex to the communication
Search/Exam
Nov 29, 2017
Communication from the Examining Division
Search/Exam
Oct 27, 2017
Examination started
Search/Exam
Jun 23, 2017
Jun 14, 2017
(Electronic) Receipt
Search/Exam
Jun 14, 2017
Letter accompanying subsequently filed items
Search/Exam
Jun 14, 2017
Letter concerning the inventor
Search/Exam
Jun 14, 2017
Letter concerning the inventor
Search/Exam
Jun 14, 2017
Letter concerning the inventor
Search/Exam
Apr 11, 2017
(Electronic) Receipt
Search/Exam
Apr 11, 2017
Apr 11, 2017
Amended claims with annotations
Search/Exam
Apr 11, 2017
Amendments received before examination
Search/Exam
Apr 11, 2017
Letter accompanying subsequently filed items
Search/Exam
Mar 13, 2017
Feb 8, 2017
Notification of forthcoming publication
Search/Exam
Feb 8, 2017
Refund of fees
Search/Exam
Feb 3, 2017
Feb 3, 2017
European search opinion
Search/Exam
Feb 3, 2017
European search report
Search/Exam
Feb 3, 2017
Information on Search Strategy
Search/Exam
Jan 20, 2017
Search started
Search/Exam
Oct 17, 2016
Communication to designated inventor
Search/Exam
Oct 17, 2016
Communication to designated inventor
Search/Exam
Oct 17, 2016
Communication to designated inventor
Search/Exam
Oct 17, 2016
Communication to designated inventor
Search/Exam
Aug 11, 2016
Abstract
Search/Exam
Aug 11, 2016
Aug 11, 2016
Claims
Search/Exam
Aug 11, 2016
Converted Sequence Listing
Search/Exam
Aug 11, 2016
Description
Search/Exam
Aug 11, 2016
Designation of inventor
Search/Exam
Aug 11, 2016
Drawings
Search/Exam
Aug 11, 2016
Matter concerning the application
Search/Exam
Aug 11, 2016
Request for grant of a European patent
Search/Exam
Aug 11, 2016
Submission concerning representation
Search/Exam
Jul 2, 2014
Jul 2, 2014
Jul 2, 2014
Jul 2, 2014