Chemically Modified Guide Rnas For Crispr/Cas-Mediated Gene Regulation
Patent No. EP3280803 (titled "Chemically Modified Guide Rnas For Crispr/Cas-Mediated Gene Regulation") was filed by Agilent on Apr 5, 2016. The application was issued on May 26, 2021.
Patent Summary
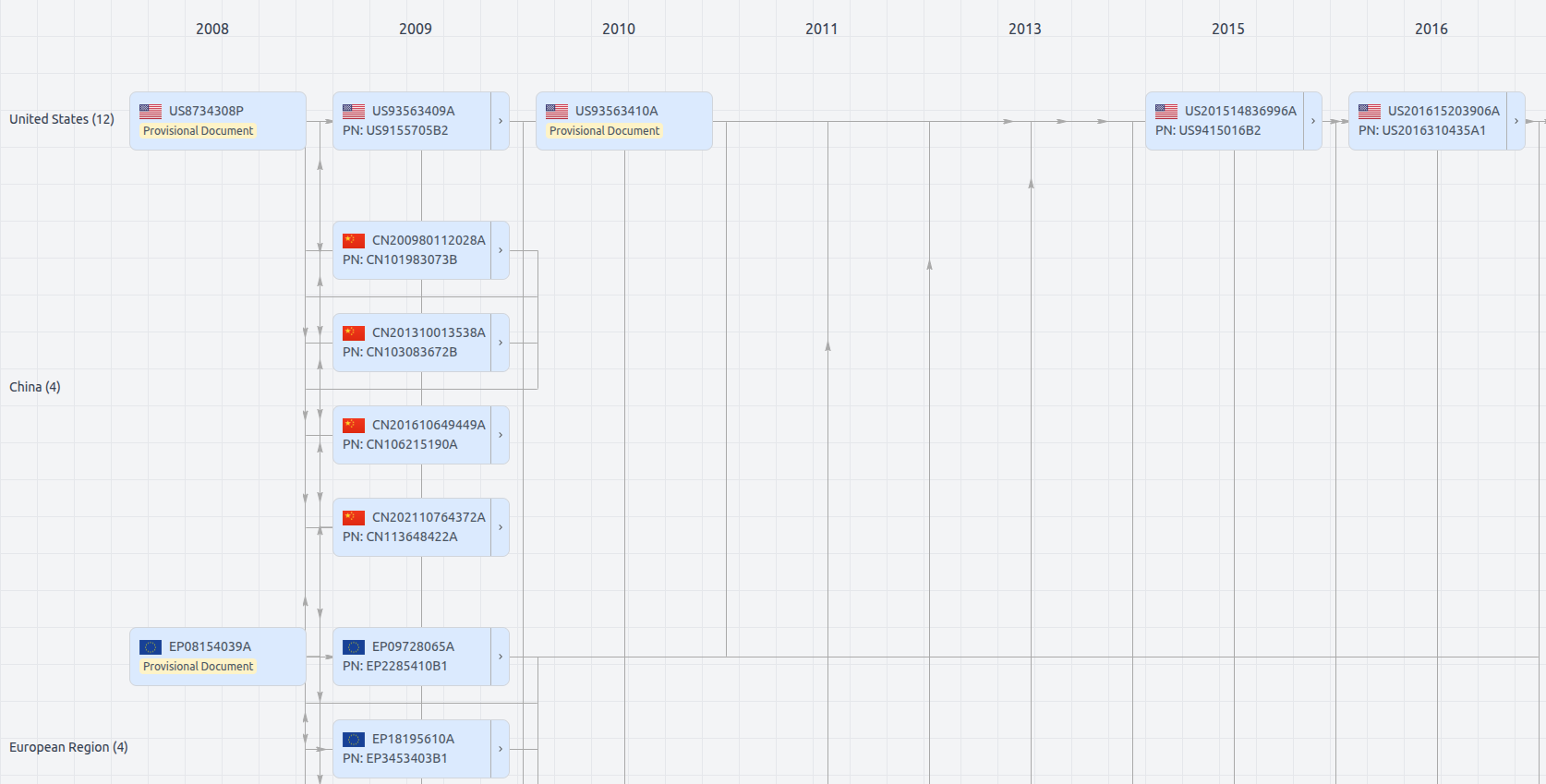
Patent Family
Patent Oppositions (3)
Patent oppositions filed by competitors challenge the validity of a granted patent. These oppositions are typically based on claims of prior art, lack of novelty, or non-obviousness. They are a key part of the process for determining a patent's strength and enforceability.
| Company | Opposition Date | Representative |
|---|---|---|
| MAIWALD | Feb 28, 2022 | MAIWALD |
| JAMES POOLE | Aug 9, 2021 | CARPMAELS & RANSFORD |
| WUESTHOFF & WUESTHOFF PATENTANWALTE PARTG MBB | Jul 28, 2021 | WICHMANN |
Dossier Documents
The dossier documents provide a comprehensive record of the patent's prosecution history - including filings, correspondence, and decisions made by patent offices - and are crucial for understanding the patent's legal journey and any challenges it may have faced during examination.
Date
Description
Get instant alerts for new documents
Feb 25, 2026
Feb 19, 2026
Feb 19, 2026
Feb 19, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Feb 18, 2026
Jan 27, 2026
Jan 27, 2026
Jan 27, 2026
Jan 22, 2026
Jan 22, 2026
Jan 8, 2026
Jan 8, 2026
Jan 8, 2026
Jan 8, 2026
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 23, 2025
Dec 2, 2025
Dec 2, 2025
Dec 2, 2025
Dec 2, 2025
Dec 2, 2025
Dec 2, 2025
Nov 27, 2025
Nov 27, 2025
Nov 27, 2025
Nov 26, 2025
Nov 26, 2025
Nov 26, 2025
Aug 20, 2025
Aug 20, 2025
Aug 20, 2025
Aug 20, 2025
Aug 20, 2025
Aug 20, 2025
Aug 20, 2025
Aug 13, 2025
Aug 13, 2025
Aug 13, 2025
Aug 13, 2025
Aug 8, 2025
Aug 8, 2025
Aug 8, 2025
Aug 5, 2025
Aug 5, 2025
Aug 5, 2025
Aug 1, 2025
Aug 1, 2025
Aug 1, 2025
Aug 1, 2025
Aug 1, 2025
Aug 1, 2025
May 2, 2024
May 2, 2024
May 2, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Apr 26, 2024
Jul 26, 2023
Jul 26, 2023
Jul 26, 2023
Jul 21, 2023
Jul 21, 2023
Jul 21, 2023
Apr 14, 2023
Apr 14, 2023
Apr 14, 2023
Apr 11, 2023
Apr 11, 2023
Apr 11, 2023
Apr 11, 2023
Jan 13, 2023
Dec 19, 2022
Dec 19, 2022
Oct 27, 2022
Oct 21, 2022
Oct 21, 2022
Oct 21, 2022
Oct 21, 2022
Oct 13, 2022
Oct 7, 2022
Oct 7, 2022
Oct 7, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Oct 4, 2022
Jun 17, 2022
Jun 17, 2022
Jun 17, 2022
Jun 13, 2022
Jun 13, 2022
Jun 13, 2022
Jun 8, 2022
Jun 8, 2022
Jun 8, 2022
Mar 23, 2022
Mar 23, 2022
Mar 23, 2022
Mar 23, 2022
Mar 4, 2022
Mar 3, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 28, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Feb 25, 2022
Aug 13, 2021
Aug 9, 2021
Aug 9, 2021
Aug 9, 2021
Aug 3, 2021
Jul 28, 2021
Jul 28, 2021
Jul 28, 2021
Jul 28, 2021
Jul 28, 2021
Jun 9, 2021
Apr 30, 2021
Apr 6, 2021
Apr 6, 2021
Apr 6, 2021
Apr 6, 2021
Dec 11, 2020
Dec 11, 2020
Dec 11, 2020
Dec 11, 2020
Dec 11, 2020
Dec 11, 2020
Oct 22, 2020
Oct 22, 2020
Oct 22, 2020
Aug 7, 2020
Aug 7, 2020
Aug 7, 2020
Aug 7, 2020
Aug 7, 2020
Jul 7, 2020
Jul 7, 2020
Jul 7, 2020
Jul 7, 2020
Jul 7, 2020
Jul 7, 2020
May 13, 2020
May 13, 2020
Apr 28, 2020
Apr 28, 2020
Apr 28, 2020
Apr 28, 2020
Apr 28, 2020
Mar 2, 2020
Mar 2, 2020
Feb 18, 2020
Oct 14, 2019
Oct 14, 2019
Sep 24, 2019
Sep 24, 2019
Sep 24, 2019
Sep 24, 2019
Sep 24, 2019
Aug 14, 2019
Aug 7, 2019
Jul 31, 2019
Jul 23, 2019
Mar 15, 2019
Mar 15, 2019
Mar 4, 2019
Mar 4, 2019
Mar 4, 2019
Oct 23, 2018
Oct 23, 2018
Jul 10, 2018
Jul 10, 2018
Jul 4, 2018
Feb 2, 2018
Feb 2, 2018
Feb 2, 2018
Jan 17, 2018
Jan 10, 2018
Jan 10, 2018
Jan 10, 2018
Dec 27, 2017
Dec 22, 2017
Dec 21, 2017
Dec 21, 2017
Dec 13, 2017
Nov 16, 2017
Nov 14, 2017
Nov 14, 2017
Nov 14, 2017
Nov 14, 2017
Nov 14, 2017
Nov 14, 2017
Nov 14, 2017
Nov 14, 2017
Nov 6, 2017
Nov 6, 2017
Nov 6, 2017
Nov 6, 2017
Oct 25, 2017
Aug 18, 2017
Oct 31, 2016
Oct 31, 2016
Oct 13, 2016
Oct 13, 2016
Jul 22, 2016
Jul 21, 2016
Jul 21, 2016
Jul 21, 2016
Jun 24, 2016
Jun 22, 2016
Jun 22, 2016
Jun 22, 2016
Jun 21, 2016
Jun 21, 2016
Jun 21, 2016
May 23, 2016
May 23, 2016
May 18, 2016
Apr 22, 2016
Apr 22, 2016
Apr 22, 2016
Apr 22, 2016
Apr 22, 2016
Apr 22, 2016
Apr 22, 2016
Apr 22, 2016
Apr 22, 2016
Apr 22, 2016
EP3280803
- Application Number
- EP16720220A
- Filing Date
- Apr 5, 2016
- Status
- Granted And Under Opposition
- Apr 23, 2021
- Publication Date
- May 26, 2021
- External Links
- Slate, Register , Google Patents