The weekly litigation news digest is live. Subscribe now
Determination Of Base Modifications Of Nucleic Acids - EP4365307
The patent EP4365307 was granted to The Chinese University OF Hong Kong on Jan 8, 2025. The application was filed on Aug 17, 2020 under application number EP24151720A. The patent is currently recorded with a legal status of "Granted And Under Opposition".
EP4365307
- Application Number
- EP24151720A
- Filing Date
- Aug 17, 2020
- Status
- Granted And Under Opposition
- Nov 1, 2024
- Publication Date
- Jan 8, 2025
- External Links
- Slate, Register, Google Patents
Patent Summary
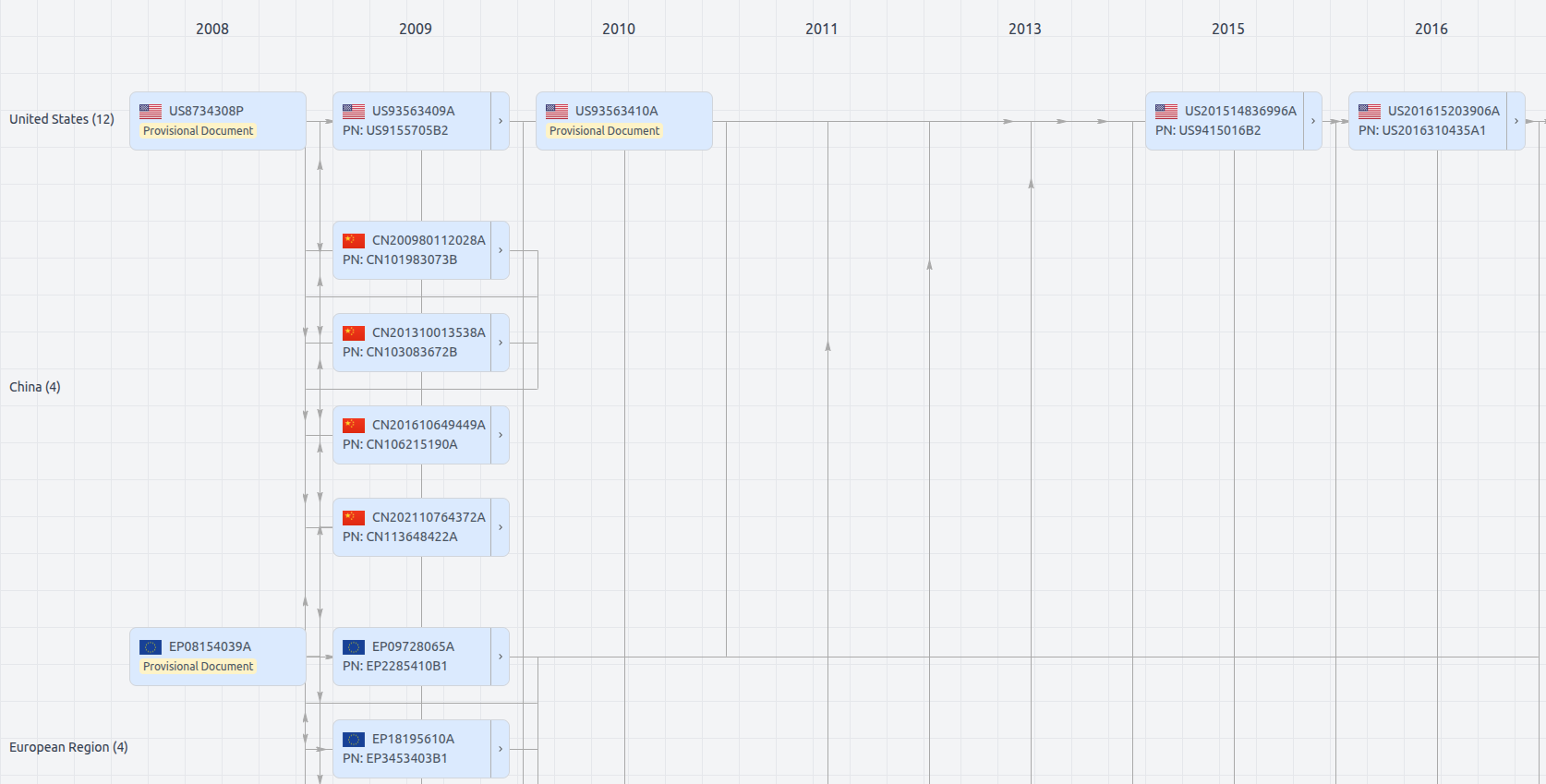
Patent Family
Patent Oppositions
Patent oppositions filed by competitors challenge the validity of a granted patent. These oppositions are typically based on claims of prior art, lack of novelty, or non-obviousness. They are a key part of the process for determining a patent's strength and enforceability.
Patent Citations (3) New
Patent citations refer to prior patents cited during different phases such as opposition or international search.
Dossier Documents
The dossier documents provide a comprehensive record of the patent's prosecution history - including filings, correspondence, and decisions made by patent offices - and are crucial for understanding the patent's legal journey and any challenges it may have faced during examination.
Date
Description
Get instant alerts for new documents