Barcoded Beads And Method For Making The Same By Split-Pool Synthesis
Patent No. US11692214 (titled "Barcoded Beads And Method For Making The Same By Split-Pool Synthesis") was filed by Roche Sequencing Solutions Inc on Dec 21, 2022.
What is this patent about?
’214 is related to the field of molecular tagging and detection , specifically for identifying and quantifying molecules within individual cells or on particles. The background acknowledges the need for improved methods to analyze gene expression and protein variants at the single-cell level, overcoming limitations of existing techniques that require large sample sizes or lack cell-specific information. The invention aims to address the challenges in multiplexed protein measurement technologies.
The underlying idea behind ’214 is to create a system where each molecule of interest is tagged with a unique identifier that reveals both its identity and its cellular origin. This is achieved by attaching a unique binding agent (UBA) to the target molecule, then linking that UBA to an epitope-specific barcode (ESB) and a cell origination barcode (COB) . The COB is constructed through a split-pool synthesis approach, ensuring each cell receives a unique barcode.
The claims of ’214 focus on a population of beads, where each bead is associated with a nucleic acid tag. This tag includes a cell origination barcode and a degenerate sequence . The key aspect is that different beads have different cell origination barcodes, allowing for the identification of the origin of the molecules attached to the beads. The method claim covers the process of adding these tags to beads using a split-pool barcoding process.
In practice, the invention involves several steps. First, target molecules within cells are bound by UBAs. Next, ESBs are linked to these UBAs. Then, a split-pool synthesis is performed to attach COBs to the ESBs, effectively creating a unique identifier for each target molecule within each cell. Finally, the tagged molecules are detected and quantified, revealing the expression levels of different targets in individual cells.
This approach differs from prior solutions by enabling highly multiplexed analysis at the single-cell level without requiring physical isolation of cells during the tagging process. The use of a split-pool synthesis to generate COBs allows for a large number of unique barcodes to be created, ensuring that each cell is uniquely identified. The addition of a degenerate sequence further enhances the uniqueness and complexity of the tags, improving the accuracy and sensitivity of the analysis.
How does this patent fit in bigger picture?
Technical landscape at the time
In the early 2010s when ’214 was filed, single-cell analysis was an emerging field, at a time when multiplexed protein measurement technologies faced significant challenges due to the complexities inherent in protein samples. Methods for accurate and sensitive detection, identification, and quantification of target molecules in every cell of a complex cell population, while retaining cell-specific information, were still needed. Existing methods often required significant amounts of biological samples or did not provide cell-specific information.
Novelty and Inventive Step
The examiner approved the claims because the prior art before January 31, 2011, did not teach or suggest beads comprising a cell origination barcode and a degenerate sequence (e.g., a random barcode), where each bead has a different cell origination barcode. While some prior art taught beads with barcodes, they failed to teach or suggest that each bead has a different cell origination barcode. Other prior art taught splitting pools of cells containing bead-oligo barcodes but did not teach or suggest that each bead has a different cell origination barcode.
Claims
This patent contains 26 claims, with independent claims 1 and 11. Claim 1 focuses on a population of beads with specific nucleic acid tags, while claim 11 focuses on a method for making barcoded beads. The dependent claims generally elaborate on the composition and characteristics of the beads and nucleic acid tags, as well as specific steps and conditions of the barcoding method.
Key Claim Terms New
Definitions of key terms used in the patent claims.
Litigation Cases New
US Latest litigation cases involving this patent.
Patent Family
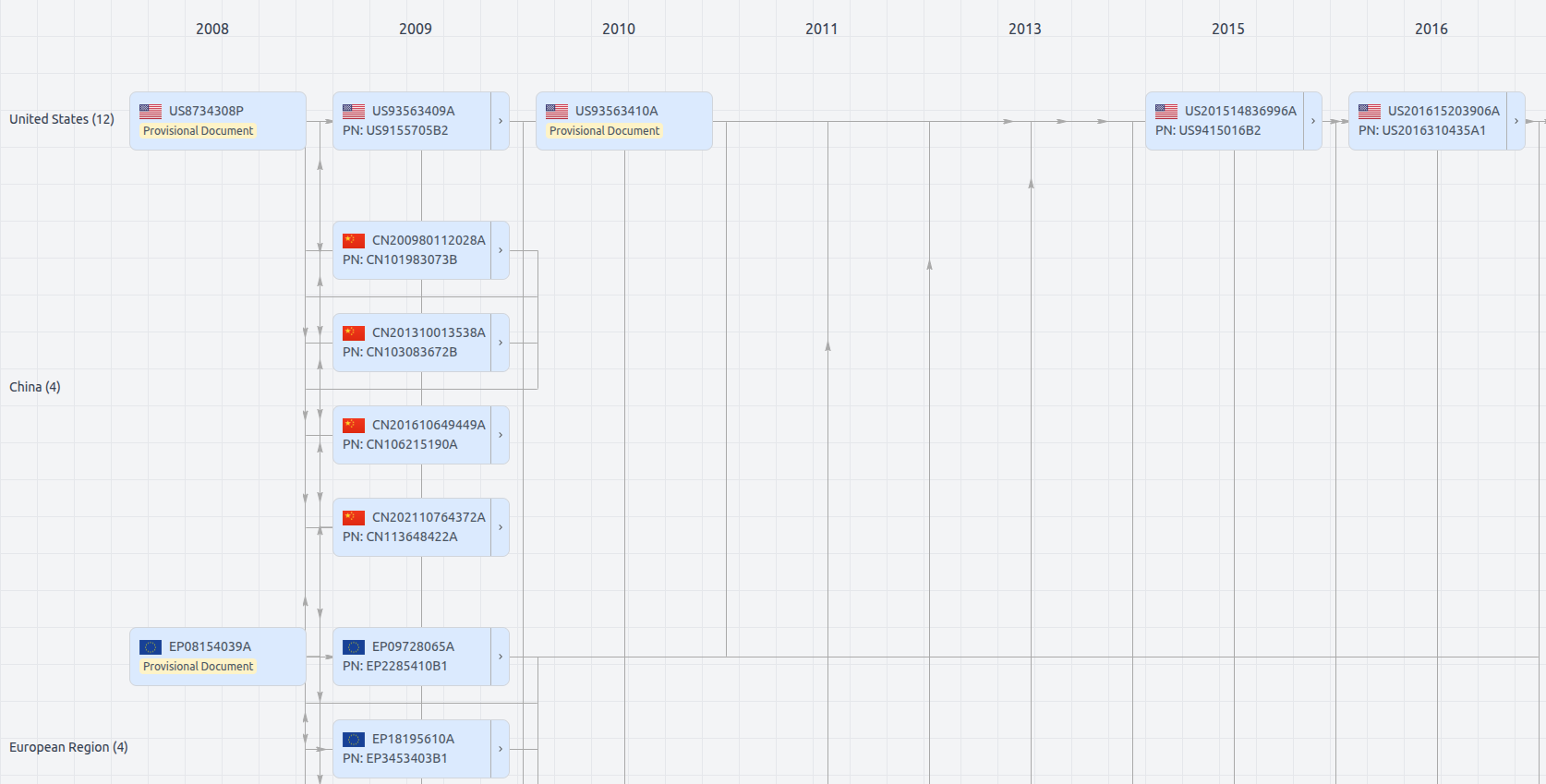
File Wrapper
The dossier documents provide a comprehensive record of the patent's prosecution history - including filings, correspondence, and decisions made by patent offices - and are crucial for understanding the patent's legal journey and any challenges it may have faced during examination.
Date
Description
Get instant alerts for new documents
US11692214
- Application Number
- US18086383
- Filing Date
- Dec 21, 2022
- Status
- Granted
- Expiry Date
- Jan 31, 2032
- External Links
- Slate, USPTO, Google Patents