Spatially Distinguished, Multiplex Nucleic Acid Analysis Of Biological Specimens
Patent No. US11739372 (titled "Spatially Distinguished, Multiplex Nucleic Acid Analysis Of Biological Specimens") was filed by 10X Genomics Sweden Ab on Feb 9, 2023.
What is this patent about?
’372 is related to the field of spatial genomics , specifically methods for analyzing the nucleic acid content of biological samples while preserving spatial information. This addresses the limitation of traditional genomic analysis, which often involves homogenizing samples, thus losing the spatial context of individual cells or regions within a tissue. The background highlights the importance of understanding tumor heterogeneity and the need for tools that can distinguish subpopulations of cancer cells.
The underlying idea behind ’372 is to spatially tag nucleic acids from a biological specimen by hybridizing them to probes with unique barcode sequences attached to a solid support. The location of each probe on the support is determined, and when target nucleic acids from the specimen hybridize to these probes, they are effectively labeled with the barcode corresponding to their location. This allows for subsequent analysis of the nucleic acids while retaining information about their original spatial context within the sample.
The claims of ’372 focus on a method comprising: (a) providing a solid support with randomly located nucleic acid probes, each containing a target capture sequence and a unique spatial tag sequence; (b) determining the spatial tag sequences at each location; (c) contacting the biological specimen with the probes; (d) hybridizing the target capture sequences to target nucleic acids from the specimen; and (e) extending the target capture sequences to produce extended probes containing complementary sequences to the target nucleic acids and the spatial tag sequences.
In practice, the invention involves creating a dense array of probes on a surface, such as a modified Illumina flow cell or a bead array. Each probe contains a unique barcode and a sequence designed to capture specific target nucleic acids, such as mRNA (using oligo-dT sequences). After the barcode locations are mapped using sequencing or hybridization, a tissue section is placed on the array. mRNA released from the tissue hybridizes to the probes, and a reverse transcription step extends the probes, incorporating the mRNA sequence alongside the spatial barcode. The tissue is then removed, and the extended probes, now spatially tagged cDNA, can be analyzed.
This approach differentiates itself from prior methods by directly linking spatial information to nucleic acid sequences at a high density. Unlike traditional methods that rely on physical separation or microdissection, ’372 uses a random array of barcoded probes to capture and tag nucleic acids in situ. This allows for a more comprehensive and spatially resolved analysis of complex biological samples, enabling researchers to study gene expression patterns and genomic variations within specific regions of a tissue or within individual cells.
How does this patent fit in bigger picture?
Technical landscape at the time
In the mid-2010s when ’372 was filed, nucleic acid analysis was typically performed using microarrays or next-generation sequencing platforms. At a time when systems commonly relied on homogenates derived from mixtures of many different cells, preserving spatial information during multiplex nucleic acid analyses was non-trivial. Hardware or software constraints made it difficult to distinguish where any particular nucleic acid resided in the biological specimen.
Novelty and Inventive Step
Claims 2-31 were rejected for obviousness-type double patenting over U.S. Patent Nos. 11,390,912, 11,299,774, 11,162,132, and 10,774,374, and pending U.S. Application No. 17/011,923. The claims were considered free of prior art. The prosecution record does NOT describe the technical reasoning or specific claim changes that led to allowance.
Claims
There are 30 claims in total, with claim 1 being the only independent claim. Independent claim 1 is directed to a method for spatially tagging target nucleic acids of a biological specimen using a solid support with randomly located nucleic acid probes. The dependent claims generally elaborate on and refine the method of independent claim 1, providing details regarding amplification, solid support characteristics, sequencing methods, and biological specimen handling.
Key Claim Terms New
Definitions of key terms used in the patent claims.
Litigation Cases New
US Latest litigation cases involving this patent.
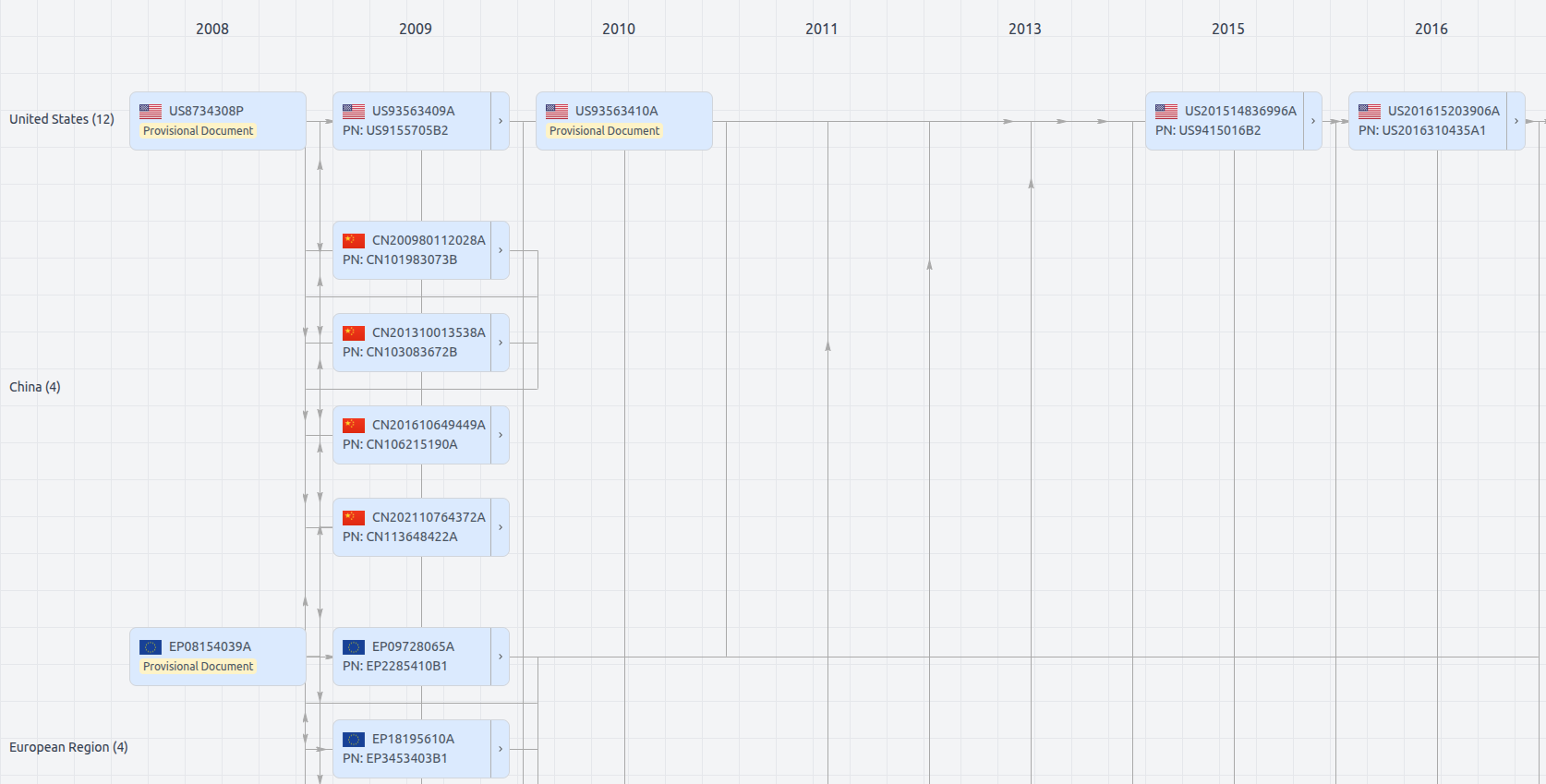
Patent Family
File Wrapper
The dossier documents provide a comprehensive record of the patent's prosecution history - including filings, correspondence, and decisions made by patent offices - and are crucial for understanding the patent's legal journey and any challenges it may have faced during examination.
Date
Description
Get instant alerts for new documents
US11739372
- Application Number
- US18166853
- Filing Date
- Feb 9, 2023
- Status
- Granted
- Expiry Date
- Apr 10, 2035
- External Links
- Slate, USPTO, Google Patents