Spatially Encoded Biological Assays
Patent No. US12297487 (titled "Spatially Encoded Biological Assays") was filed by Prognosys Biosciences Inc on Dec 6, 2024.
What is this patent about?
’487 is related to the field of biological assays, specifically those used to determine the spatial distribution of biological molecules within a sample. Traditional methods like in situ hybridization and microarrays lack the ability to simultaneously measure the expression of many genes or the presence/activity of multiple proteins at many spatial locations. This patent addresses the need for reproducible, high-resolution spatial maps of biological molecules in tissues.
The underlying idea behind ’487 is to create a system where biological targets within a sample are tagged with location-specific identifiers, allowing for their detection and quantification while preserving spatial context. This is achieved by delivering encoded probes to the sample in defined spatial patterns. Each probe interacts with a specific biological target and carries a coding tag that identifies the location where the interaction occurred. After the assay, the probes are pooled, and the coding tags are read out, allowing the abundance of each target to be mapped back to its original location.
The claims of ’487 focus on a composition comprising a substrate, a tissue section disposed on the substrate, and a plurality of capture probes. Each capture probe has three key components: a target-binding domain (a nucleic acid sequence that binds to a target nucleic acid), a nucleic acid sequence that identifies the probe's location on the substrate, and a primer binding site. Claim 17 focuses on a similar composition within a flow cell, where the capture probes are immobilized in wells.
In practice, the system works by first affixing a biological sample, such as a tissue section, to a substrate. Then, encoded probes are delivered to specific locations on the sample. These probes are designed to bind to target molecules of interest, such as specific RNA transcripts or proteins. After allowing the probes to interact with their targets, unbound probes are washed away. The remaining probes, now bound to their targets and carrying location-specific tags, are then processed for sequencing.
The key differentiation from prior approaches lies in the combination of spatial encoding and high-throughput sequencing. Traditional methods either lack the ability to analyze many targets simultaneously or lose spatial information during the analysis. ’487 overcomes these limitations by using a combinatorial encoding scheme where location is encoded by a combination of tags, significantly reducing the number of unique probes needed. The use of next-generation sequencing allows for the simultaneous readout of millions of tags, enabling the creation of high-resolution spatial maps of biological activity.
How does this patent fit in bigger picture?
Technical landscape at the time
In the early 2010s when ’487 was filed, high-throughput gene expression analysis and protein analysis were established techniques, at a time when microarrays, qPCR, and in situ PCR were commonly used for transcriptomic analysis. However, simultaneous measurement of many genes or proteins at many spatial locations in a sample was non-trivial, when systems commonly relied on methods with limitations in multiplexing, spatial resolution, or scalability.
Novelty and Inventive Step
This document does not contain examiner reasoning or explanations relevant to allowance.
Claims
US12,297,487 has 29 claims, of which claims 1 and 17 are independent. The independent claims focus on compositions comprising a substrate with capture probes and a tissue section, or a flow cell with capture probes, for spatial analysis. The dependent claims generally specify details and variations of the compositions described in the independent claims.
Key Claim Terms New
Definitions of key terms used in the patent claims.
Litigation Cases New
US Latest litigation cases involving this patent.
Patent Family
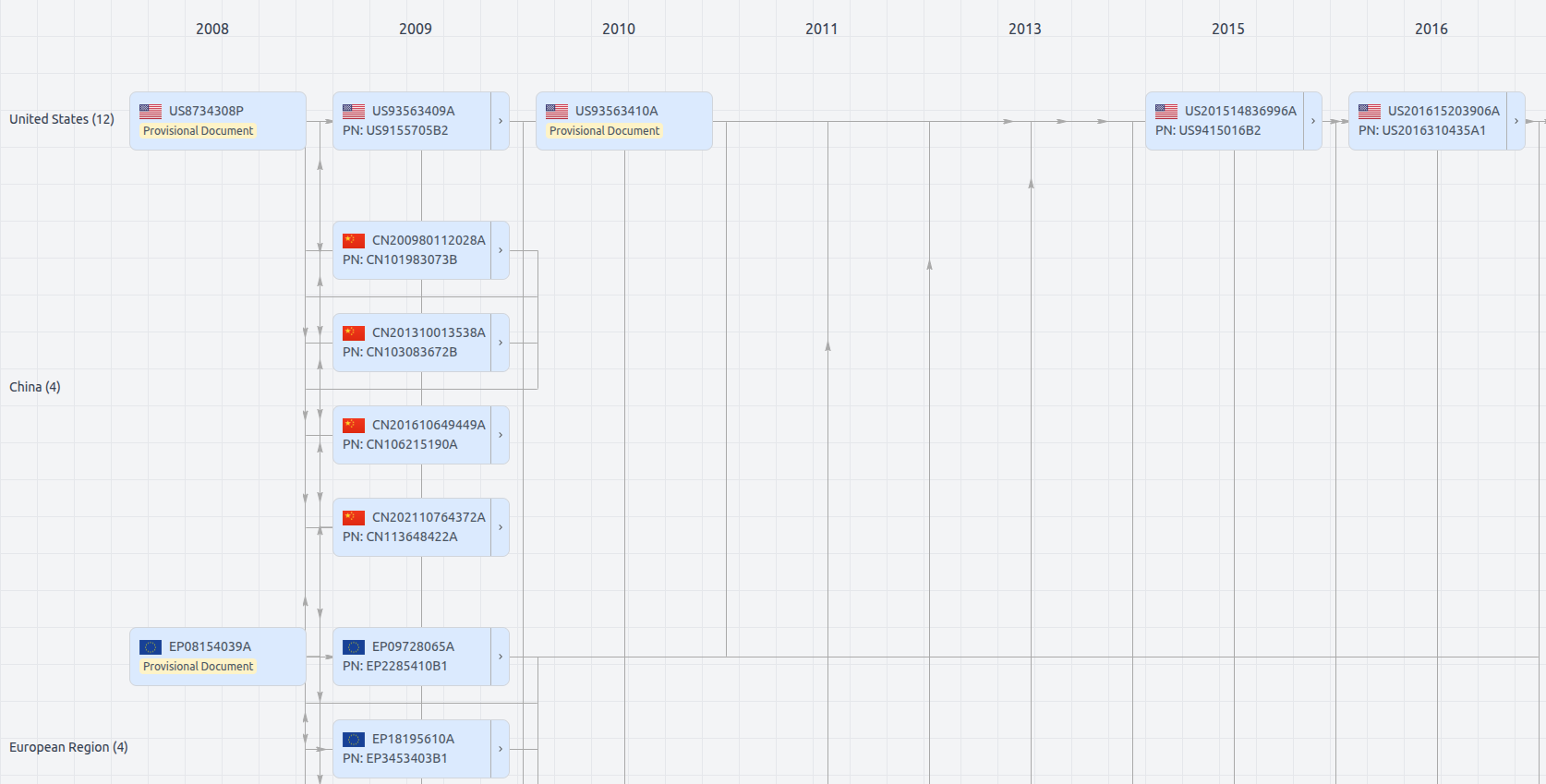
File Wrapper
The dossier documents provide a comprehensive record of the patent's prosecution history - including filings, correspondence, and decisions made by patent offices - and are crucial for understanding the patent's legal journey and any challenges it may have faced during examination.
Date
Description
Get instant alerts for new documents
US12297487
- Application Number
- US18972052
- Filing Date
- Dec 6, 2024
- Status
- Granted
- Expiry Date
- Apr 5, 2031
- External Links
- Slate, USPTO, Google Patents